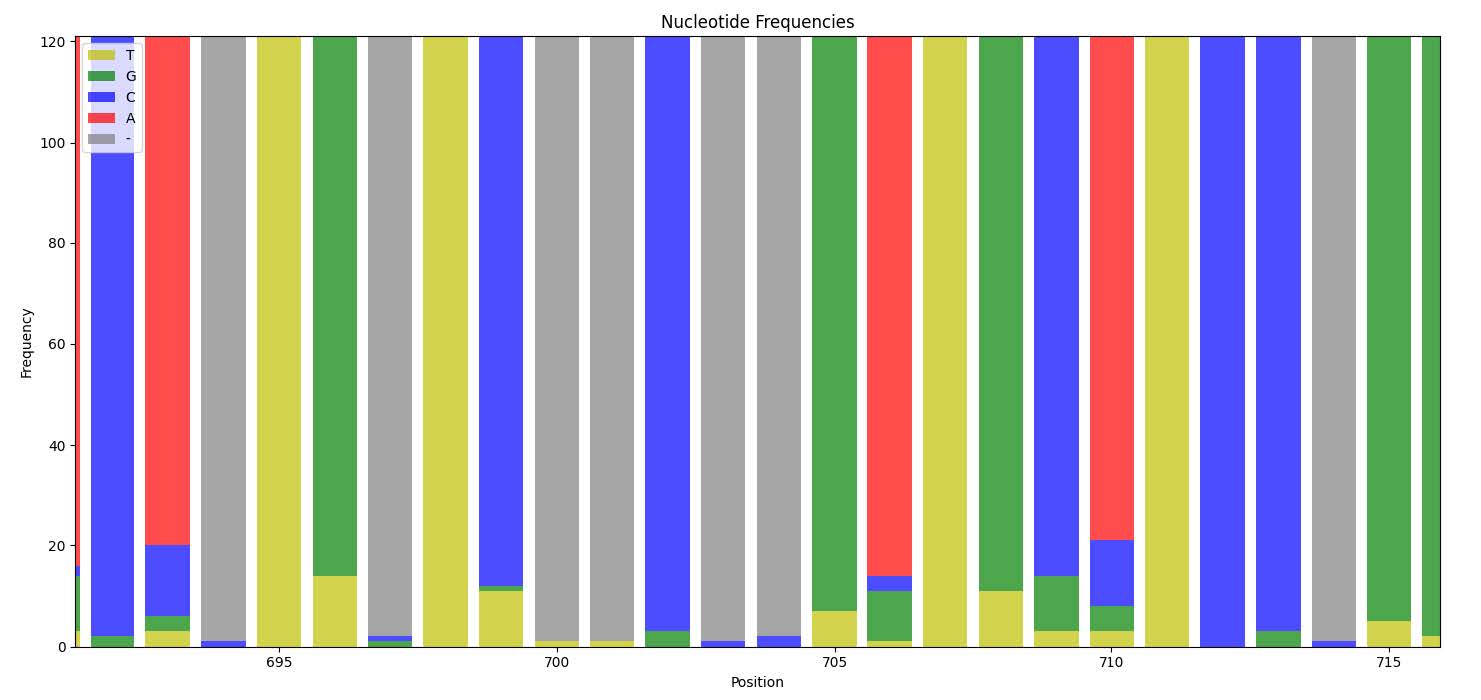
Normally, a consensus version of a gene is called in a single sample and the rest are ignored. With many organisms being only 1-100x covered in microbiomes, it is not feasible to distinguish between sequencing errors and real, rare variants. However, if we recruit reads from hundreds of thousands of samples to build very deep alignments over each gene, we can subtract the background noise of sequencing errors and start to study the rate at which each position mutates either globally or in specific subsets of the public microbiome samples.
In our new project, by using metagenomic samples from all around the world, we want to focus on specific genes of interest and quantify the rate at which mutations occur. In this brief presentation, I present the first steps on Million Miles High project.
Nikiforos Pyrounakis’ presentation