Antimicrobial resistance is a huge burden on society causing many deaths and loss of workforce productivity due to untreatable or hard-to-treat infections by resistant pathogens. One of the most used class of antibiotics, namely the beta-lactams (includes penicillin), are made inert or ineffective by proteins/enzymes called beta-lactamases. Currently, it is estimated that almost 3000 unique proteins encoding beta-lactamases have been identified. This huge diversity of resistance mechanisms each provide a slightly different type of protection toward the different classes of beta-lactam drugs (e.g. penicillin, carbapenems, cephalosporins, monobactams, etc.).
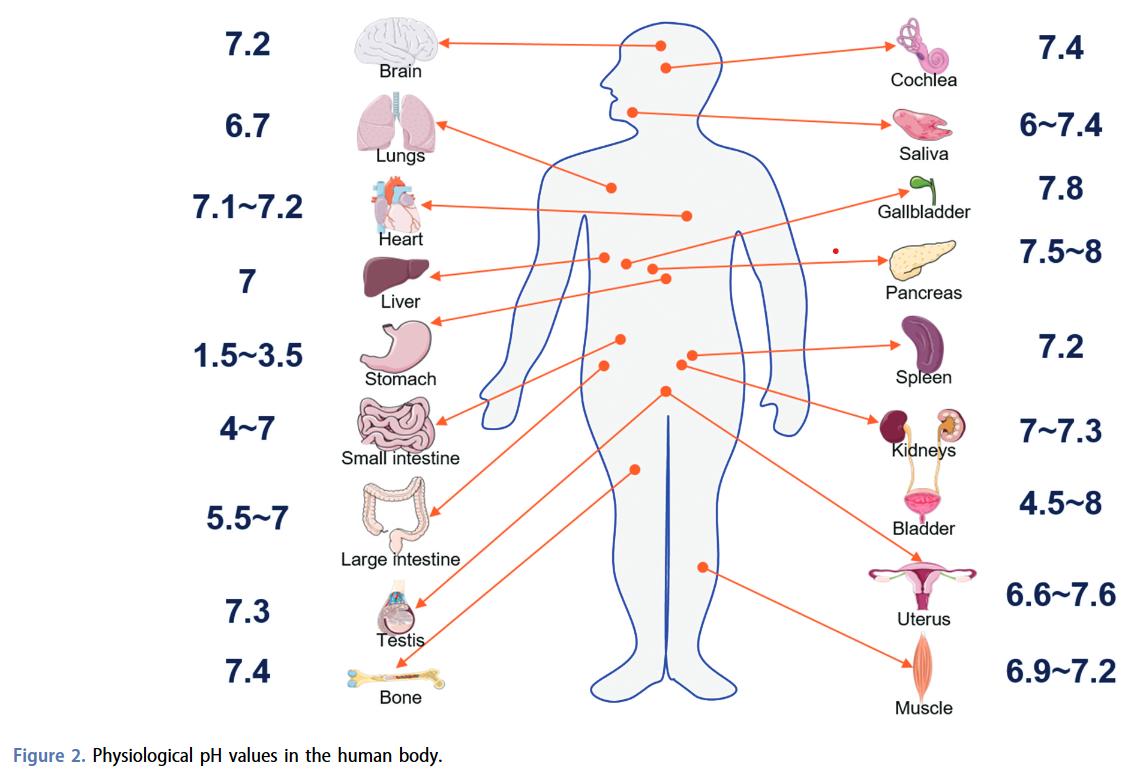
We also know from virtually every other enzyme that the environmental conditions, such as temperature and pH, are critical for their function. However, this is a completely overlooked aspect of antimicrobial susceptibility testing, which uses the same standardized media, temperature and pH. While this leads to highly reproducible results, we have preliminary data, which suggests that the environmental conditions are as important to beta-lactamase activity as it is to other enzymes. Failing to account for this could in a worst-case scenario lead to antibiotic treatment failure, where a seemingly susceptible bacterial infection is resistant in certain environments.
With this project, we wish to answer the following questions:
- To what degree is the activity of beta-lactamases impacted by changes in pH or temperature?
- Are these effects dependent on the specific beta-lactam antibiotic (i.e., the enzyme substrate)?
Another unanswered question relating to antibiotic resistance that we wish to address is why bacteria obtain and maintain multiple different genes encoding beta-lactamases with, seemingly, the same function.
- Will bacteria with specific combinations of beta-lactamases prevail under specific and especially changing environmental conditions (i.e., a change in infection site or host, which can change pH and or temperature)?
To answer these questions, we will construct isogenic laboratory strains each expressing different beta-lactamases under highly controlled transcriptional and translational control to facilitate the best possible comparisons. These strains will then be tested for antimicrobial susceptibility under different environmental conditions.
Mikkel Anbo’s presentation
Reference for graphic at top of the page:
Gaohua, L., et al. "Crosstalk of physiological pH and chemical pKa under the umbrella of physiologically based pharmacokinetic modeling of drug absorption, distribution, metabolism, excretion, and toxicity." Expert opinion on drug metabolism & toxicology 17.9 (2021): 1103-1124.