The combat against antibiotic resistance has never been as heated as today. We use many antibiotics around the globe, which apply evolutionary pressure on bacteria to develop new ways of resistance. Methods that help discover new resistance genes could impact the ongoing fight against infectious diseases.
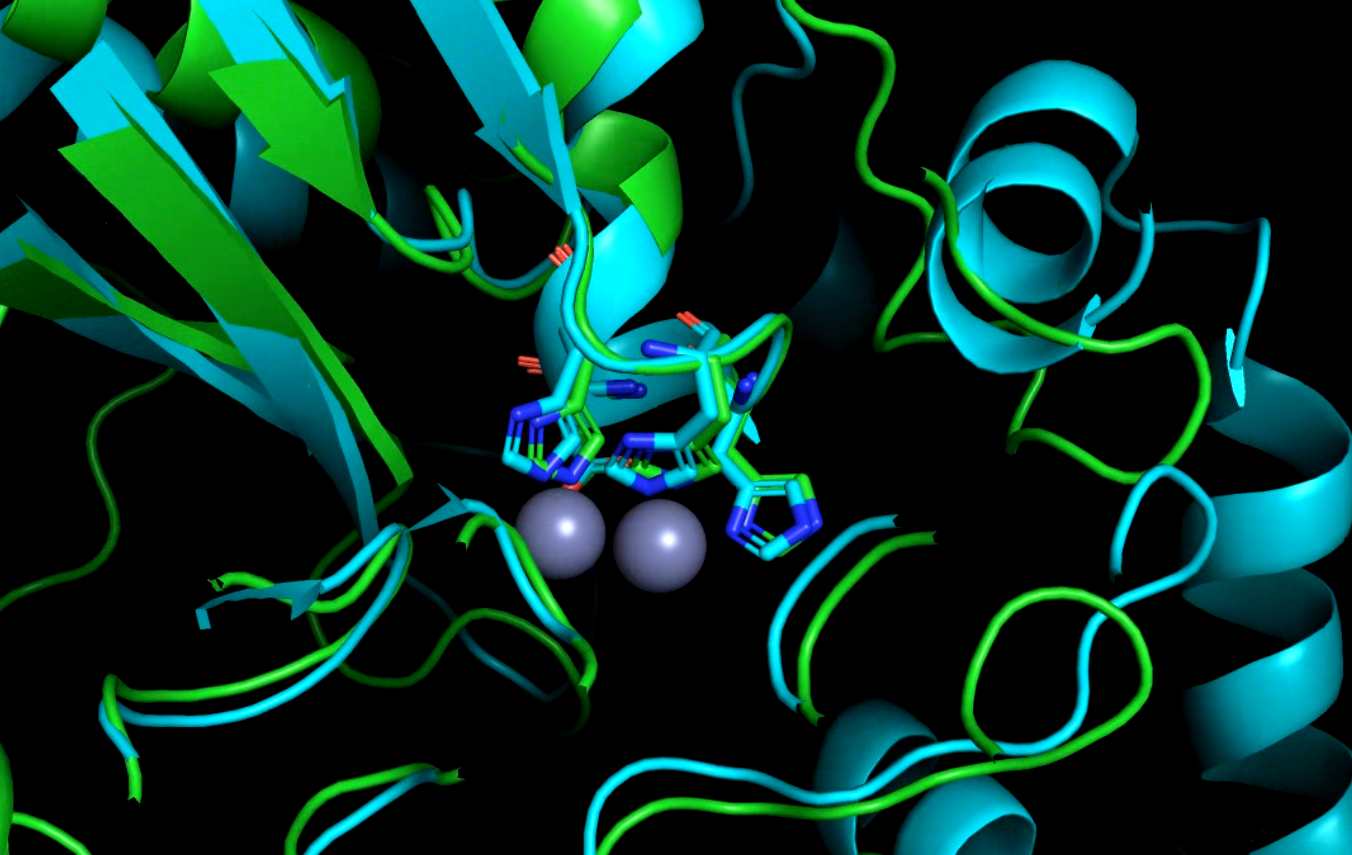
We hypothesize that sewage could be a potential reservoir of many different organisms with various resistance genes, therefore the raw data of DNA sequences is coming from a global sewage study. The first step is the identification of genes and translating them to amino acid sequences. Then, by using Hidden-Markov-Models all the sequences are aligned to known profiles of resistance genes. Then with different filters, the data was reduced to unknown, beta-lactamase-like hypothetical proteins, focusing on those which have conserved active sites. This subgroup of POI (Protein Of Interest) is roughly 0.05% of the original data.
AlphaFold2 is used for predicting the 3D structure of POI, and then by structural alignment, the promising-looking ones are checked against known 3D structures. If the evaluation methods of the 3D structure show high similarity, the upcoming step is to validate our findings in vitro.
Attila Beleon’s presentation